Our research complies with all the relevant ethical regulations. The Animal Use and Care Committee of the University of Pretoria evaluated and approved the experimental protocol and collection of all samples (ethics clearance number: NAS022/2021), with DAFF section 20 approval (SDAH-Epi-21051907211). Animal experiments at QMUL were approved by the local ethical review committee and in accordance with the Home Office Animals in Scientific Procedures Act 1986.
Animals
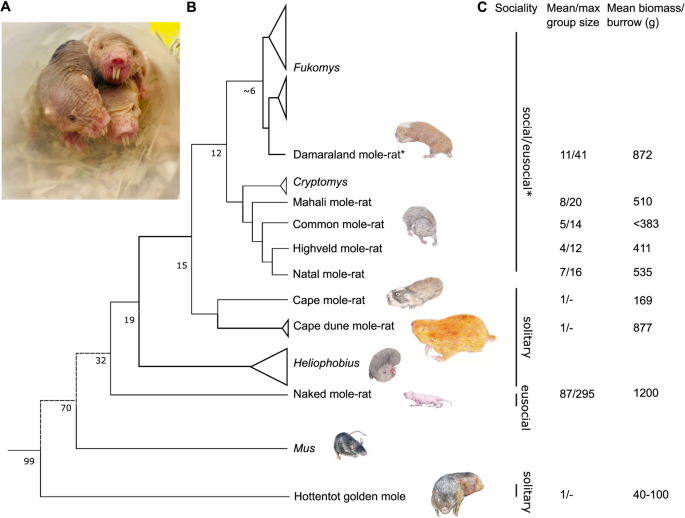
NMRs were bred at the School of Biological and Behavioural Sciences, Queen Mary University of London. The non-breeding adult NMRs used in this study were second-generation or more captive-born, descended from animals captured in Kenya in the 1980s. Colonies were maintained using artificial burrow systems (group-housed in interconnected multi-cage systems) at 30 °C and 21% O2 in 50% humidity with a 12 h light cycle. Their diet consisted of fresh vegetables, fruit, and tubers (sweet potatoes) ad libitum. All water requirements were obtained from the food resources. A noticeable drop in the heart-to-body mass ratio is observed in African mole-rats after a year in captivity. At least one year of captivity has resulted in the wild-caught and captive-bred African mole-rat species having a similar heart-to-body mass ratio. Both sexes (mole-rats) were used in the study, as summarised in Supplementary Table 1). The ages selected for this study allowed for physiological age matching such that all animals were at equivalent percentages of maximum lifespan and, therefore, not the same chronological age. C57/BL6 (J strain, male) mice were purchased from Charles River Laboratories, UK and kept in individually ventilated cages, with access to food (PicoLab Mouse Diet 20 EXT 5R58, irradiated, I-DIET-5R58-9KG-BG, IPS) and water ad libitum. All mice are housed in IVCs (Individual Ventilated Cages). Enrichment is provided in the form of tunnels and chew sticks. Cages are cleaned out under a LEV hood when required, the maximum time between cage changes is 14 days.
Georychus capensis, Bathyergus suillus, Cryptomys hottentotus hottentotus, C. h. pretoriae, C. h. mahali, and C. h. natalenesis were wild-captured in South Africa using Hickman live traps, baited with a small piece of sweet potato. All traps were monitored for captures every 2–3 hours over the course of the day and left overnight, being checked first thing in the morning. Permission to capture these species was obtained from all landowners, and a collecting permit was obtained from the relevant nature conservation authorities (Permit number: Western Cape- CN44-87-13780, Gauteng- CPF6-0124, Kwa-Zulu Natal- OP1545/2021). Fukomys damarensis were laboratory maintained. Captured animals were brought back and individually housed at the University of Pretoria at ~27 °C and 21% O2 in 50% humidity with a 12 L:12D light cycle. All animals were housed in large polyurethane crates (1 × 0.5 × 0.5 m) with wood shavings and paper towelling for nesting material in temperature-controlled rooms. All wild-caught African mole-rat species were maintained in captivity for at least one year before tissue harvesting. Due to difficulty in maintaining the Golden moles in captivity and their short lifespan, they were kept captive for ~3 weeks prior to tissue harvest. Each animal was terminally anaesthetised using isoflurane. Immediately upon cessation of breathing, heart was dissected and within 60 seconds placed in cryovial containing RNA later, and flash-frozen in liquid nitrogen. Subsequently, samples were placed in the −80 °C freezer for storage.
RNAseq/transcriptome analysis
Total RNA extraction and analysis were performed commercially by BGI Tech solutions (Hong Kong) using sequencing platform DNBseq and read length PE 100 bp. We sequenced 41 samples as follows: Naked mole-rat, Heterocephalus glaber (n = 6), Cape mole-rat, Georychus capensis (n = 5), Cape dune mole-rat, Bathyergus suillus (n = 4), Common mole-rat, Cryptomys hottentotus hottentotus, C. h. natalenesis (n = 5), Mahali mole-rat, C. h. mahali (n = 5), Highveld mole-rat C. h. pretoriae (n = 5), Damaraland mole-rat, Fukomys damarensis, Hottentot golden mole, Amblysomus hottentotus (n = 2) and C57BL6 mice Mus musculus (n = 6). An average of 4.28 Gb of sequence was generated per sample.
Standard bioinformatic processing was performed as a service by BGI Tech Solutions. Briefly, the reads mapped to rRNAs were removed to produce rawdata, then low-quality reads were filtered where more than 20% of the qualities of the base were lower than 10, reads with adaptors and reads with unknown bases (N bases >5%) were also removed to produce “clean reads”. Clean reads were mapped onto reference genome (Heterocephalus glaber Ensemble_release-108). Genome mapping was conducted using HISAT2 (Hierarchical Indexing for Spliced Alignment of Transcripts; http://www.ccb.jhu.edu/software/hisat).
The average mapping ratio with the reference genome was 18.13%, while the average mapping ratio for genes was 32.28%; 20,285 genes were identified. Mapping was followed by novel gene prediction, SNP and INDEL calling, and gene splicing detection. StringTie (http://ccb.jhu.edu/software/stringtie) was used to reconstruct transcripts, while Cuffcompare and Cufflinks tools (http://cole-trapnell-lab.github.io/cufflinks) were used to compare reconstructed transcripts to the reference annotation. Novel transcripts were defined as (i) unknown, intergenic transcripts, (ii) transfrags falling entirely within a reference intron, (iii) generic exonic overlaps with a reference transcript, and (iv) potentially novel isoforms (fragments) where at least one splice junction is shared with a reference transcript. CPC was then used to predict the coding potential of novel transcripts. Novel coding transcripts were then merged with reference transcripts to get a complete reference, and downstream analysis was based on this complete reference. GATK33 was deployed to call SNPs and INDELs for each sample.
Finally, DEGs were identified between samples with DEseq234, and clustering analysis and functional annotations performed. Hierarchical clustering for DEGs was performed using pheatmap, a function of R. Following Gene Ontology Analysis of DEGs GO functional enrichment analysis was undertaken using phyper, a function of R. The analysis pipeline is summarised in Supplementary Fig. 8.
Due to the limited mapping rate using the naked mole-rat as a reference for the comparison with the mouse C57/BL6 transcriptome (and vice versa), and the limited common expression of the most expressed genes in each species, it was not possible to do the same range of analyses for the mouse versus the naked mole-rat (or mouse versus the other species), as for the naked mole-rat versus the subterranean species listed above. Instead, as a workaround, we identified orthologous genes in the naked mole-rat and mouse in order to compare gene expression between the species. For transcripts that mapped to the naked mole-rat genome, we extracted the corresponding gene sequences directly from the naked mole-rat genome (GCA_944319725.1).
For novel transcripts, we used associated protein IDs identified by BGI and downloaded the corresponding gene sequences using the Entrez Direct Utilities35. We used the Basic Local Alignment Search Tool (BLAST; v2.11.036) with parameters -evalue 1e-50 -perc_identity 80 to query all sequences against the mouse reference genome (GCF_000001635.27). We took the best hit for each transcript and calculated the log2 fold change between the mean abundance (FPKM) of each species. We plotted the data using the R package ggplot235,36.
Langendorff heart perfusion
Beating hearts were excised from terminally anaesthetised naked mole-rats (n = 5) and C57/BL6 mice (n = 5) (Charles River, UK) for Langendorff perfusions as previously described37. Ex vivo cardiac function data was recorded using ADINstruments Chart software (v6). All perfusions were carried out at 37 °C. After 20 minutes of equilibration, hearts were subject to 20 minutes of global normothermic ischaemia (37 °C)38. Hearts were snap-frozen at the end of the protocol using a Wollenberger clamp pre-cooled in liquid N2. Ischaemia/reperfusion (I/R) tissue damage in NMRs was compared to C57BL6 mouse heart. Coronary effluent was analysed using the lactate quantification kit (Sigma Aldrich Merck, UK).
The comparison between the mouse and the NMR was made as they are similar-sized small rodents with widely different tolerance of hypoxia. Furthermore, mouse is widely used due to its anatomical, physiological, and genetic similarity to humans39. We were unable to repeat ex vivo cardiac perfusion protocols in all other 5 mole-rat genera hearts as they are not kept in captive colonies in the UK. Blood samples were collected at the time of experimental endpoint into heparinized tubes, centrifuged, plasma flash frozen in liquid N2 and stored in −80 °C until analysis.
Plasma biochemical profiling was carried out by the MRC Mouse Biochemistry Laboratory (Addenbrookes NHS Hospital, Cambridge). Plasma triglycerides (4 µl sample) were measured using Siemens Dimension EXL clinical sample analyser with Siemens Healthcare reagents (DF69A). Enzymatic assay was based on lipoprotein lipase conversion of triglycerides into free glycerol and fatty acids40. Plasma glucose was quantified using the automated enzyme assay on the Siemens Dimension EXL analyser (3 µl of sample, reagent manufacturer Siemens Healthcare, DF30). Assay is an adaptation of the hexokinase-glucose-6-phosphate dehydrogenase method41.
Plasma lactate (8 µl of sample) was analysed using the automated assay (Siemens Dimension EXL analyser, Siemens Healthcare product code DF54). Plasma insulin was analysed using guinea pig Insulin (INS) ELISA (abx150418, Abbexa, USA).
Myocardial infarct size quantification
Myocardial infarct size was quantified as described previously38,42. In brief, after 20 min of equilibration, Langendorff perfused hearts (n = 3/ group) were subject to 20 min of global normothermic ischaemia and 2 hours of reperfusion. At the end of the protocol, hearts were perfused for 10 mins with 3% triphenyltetrazolium chloride (TTC) in KH Buffer followed by 10 min incubation in 3% TTC-KH. Tissue was sectioned (mouse heart gauge, Zivic instruments, USA), and infarct field was scanned using HPSmart Colour Laser Scanner Software (V14.1.0) quantified using ImageJ Software (v 1.53t)38.
1H NMR spectroscopy
Powdered heart samples were subject to methanol/water/chloroform phase extraction37. Frozen heart tissue was homogenised in 2 mL each of ice-cold methanol, chloroform, and Millipore water and vortexed. Samples were centrifuged for 1 hour at 2500 × g at 4 °C to separate aqueous, protein, and lipid layers. The upper aqueous phase was separated, 20–30 mg chelex-100 was added to chelate paramagnetic ions, vortexed, and centrifuged at 2500 × g for 1 minute at 4 °C. The supernatant was transferred to a fresh falcon tube containing 10 μL of universal pH indicator solution followed by vortexing and lyophilization. Dual-phase-extracted metabolites were reconstituted in 600 μL of deuterium oxide [containing 8 g/L NaCl, 0.2 g/L KCl, 1.15 g/L Na2HPO4, 0.2 g/L KH2PO4 and 0.0075% w/v trimethylsilyl propanoic acid (TSP)] and adjusted to pH ≈ 6.5 by titrating with 100 mM hydrochloric acid. Coronary effluents were frozen immediately upon collection in N2 and stored at –80 C until the NMR analysis. 100 μl of D2O containing 0.0075% w/v trimethylsilyl propanoic acid (TSP) as internal reference was added to 500 μl of effluent.
1H nuclear magnetic resonance spectra were acquired using a vertical-bore, ultra-shielded Bruker 14.1. tesla (600 MHz) spectrometer with a bbo probe at 298 K using the Bruker noesygppr1d pulse sequence. Acquisition parameters were 128 scans, 4 dummy scans and 20.8 ppm sweep width, acquisition time of 2.6 s, pre-scan delay of 4 s, 90° flip angle, and experiment duration of 14.4 minutes per sample. TopSpin (version 4.0.5) software was used for data acquisition and for metabolite quantification. FIDs were multiplied by a line broadening factor of 0.3 Hz, and Fourier-transformed, phase and automatic baseline-correction were applied. Chemical shifts were normalised by setting the TSP signal to 0 ppm. Metabolite peaks of interest were initially integrated automatically using a pre-written integration region text file and then manually adjusted where required. Assignment of metabolites to their respective peaks was carried out based on previously obtained in-house data, confirmed by chemical shift and using Chenomx NMR Profiler Version 8.1 (Chenomx, Canada). Peak areas were normalised to the total metabolite peak area. Quantification of glycogen was performed by 1H magnetic resonance, giving a measure of the concentration of glucose monomers that are present in the observed peak. Being a large macromolecule, with possible differences in the mobility of glycosyl units, glycogen has been reported to be fully visible by MRS. The modified dual-phase Folch extraction method used for separating aqueous and lipid metabolites was not optimised for the extraction of glycogen, however, all samples underwent the same extraction procedure allowing between species comparison.
Western blotting
Powdered heart samples were homogenised (100 µL of buffer per 10 mg of cardiac tissue) on ice in 100 mM Tris buffer (pH 7.4) supplemented with complete mini EDTA-free protease inhibitor (Roche) using a glass tissue grinder. The tissue homogenates were re-suspended in an equal volume of 2× reducing SDS sample buffer. Proteins were resolved by SDS-PAGE (4–20% Mini-PROTEAN TGX, Bio-Rad or Novex 4–20% Tris-Glycine, ThermoFisher Scientific) using Mini-Protean 3 system (Bio-Rad) or XCell4 SureLock Midi Cell and transferred using semi-dry system (Bio-Rad) to 22 μm PVDF or Nitrocellulose membranes (Bio-Rad). Following the transfer, membranes were blocked for one hour with 5 % non-fat dried milk in TPBS at RT.
After blocking, membranes were immunoprobed with primary antibodies dissolved in 5 % non-fat dried milk in TPBS overnight. Subsequently, membranes were washed three times with TPBS and incubated with IR-dye conjugated secondary antibodies in Intercept blocking buffer (LI-COR) or with horseradish peroxidase-coupled anti-rabbit IgG antibody in 5 % non-fat dried milk in TPBS for one hour at RT in the dark. After the incubation, membranes were washed three times with TPBS and imaged using Odyssey DLx infra-red imaging system (LI-COR) or ECL reagent (ThermoFisher Scientific) and imaging cabinet. Protein bands were analysed and quantified using Image Studio Lite (LI-COR) or ImageJ (NIH).
List of vendors and dilution of antibodies used in this study
Anti HIF-1α Proteintech (#20960-1-AP); (Clone developed against protein sequence including amino acids 574–799 of the human HIF-1α protein; GenBank: BC012527.2) 1:1000.
Anti HIF-1α Abcam (#ab 179483); (Clone developed against recombinant fragment. This information is proprietary to Abcam and/or its suppliers) 1:1000.
Anti HIF-1α Novus lab (#NB100-479); (Clone developed against a fusion protein including amino acids 530–825 of the mouse HIF-1α protein; Uniprot #Q61221) 1:1000.
Anti β-Tubulin ThermoFisher Scientific (#62204) 1:5000.
Anti α-Actin Merck Millipore (#MAB1501) 1:3000.
Total OXPHOS Rodent WB Antibody Cocktail (#ab110413) 1:1000.
Statistics and reproducibility
Data are presented as mean ± SEM. Comparison between groups was performed by Student’s t test (Gaussian data distribution), two-way analysis of variance (ANOVA) with Bonferroni’s correction for multiple comparison, and one-way ANOVA using Bonferroni’s correction for multiple comparisons where applicable. Normality of data distribution was examined using Shapiro–Wilk’s normality test. Statistical analysis was performed using GraphPad Prism (v9) software. Unclassified PCA was performed using a PCA toolbox in Matlab. Data was autoscaled prior to PCA calculation using venetian blinds cross-validation and 5 cv groups43. All quantified analytes were included in the analysis. Classified discriminant analysis was performed using a classification toolbox in Matlab using linear discriminant analysis, bootstrap validation, and 100 iterations. Hierarchical cluster analysis was performed in Matlab using the function clustergram. No statistical method was used to predetermine sample size, and sample sizes were governed by the availability of rare biological material used in the study. No data were excluded from the analyses; the experiments were randomised; the investigators were blinded to allocation during outcome assessment. Differences were considered significant when P < 0.05. DEGs were visualised with enhanced volcano plots implemented in R-Studio44.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.