Nayfach, S. et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 6, 960–970 (2021).
Google Scholar
Gregory, A. C. et al. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe 28, 724–740.e8 (2020).
Google Scholar
Nishijima, S. et al. Extensive gut virome variation and its associations with host and environmental factors in a population-level cohort. Nat. Commun. 13, 5252 (2022).
Google Scholar
Shah, S. A. et al. Expanding known viral diversity in the healthy infant gut. Nat. Microbiol. 8, 986–998 (2023).
Google Scholar
Camarillo-Guerrero, L. F., Almeida, A., Rangel-Pineros, G., Finn, R. D. & Lawley, T. D. Massive expansion of human gut bacteriophage diversity. Cell 184, 1098–1109.e9 (2021).
Google Scholar
Zuppi, M., Hendrickson, H. L., O’Sullivan, J. M. & Vatanen, T. Phages in the gut ecosystem. Front. Cell. Infect. Microbiol. 11, 822562 (2022).
Google Scholar
Borodovich, T., Shkoporov, A. N., Ross, R. P. & Hill, C. Phage-mediated horizontal gene transfer and its implications for the human gut microbiome. Gastroenterol. Rep. 10, goac012 (2022).
Google Scholar
Schroven, K., Aertsen, A. & Lavigne, R. Bacteriophages as drivers of bacterial virulence and their potential for biotechnological exploitation. FEMS Microbiol. Rev. 45, fuaa041 (2021).
Google Scholar
Montassier, E. et al. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat. Microbiol. 6, 1043–1054 (2021).
Google Scholar
Federici, S., Nobs, S. P. & Elinav, E. Phages and their potential to modulate the microbiome and immunity. Cell. Mol. Immunol. 18, 889–904 (2021).
Google Scholar
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Google Scholar
Shkoporov, A. N. et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26, 527–541.e5 (2019).
Google Scholar
Zuo, T. et al. Human-gut-DNA virome variations across geography, ethnicity, and urbanization. Cell Host Microbe 28, 741–751.e4 (2020).
Google Scholar
Gulyaeva, A. et al. Discovery, diversity, and functional associations of crAss-like phages in human gut metagenomes from four Dutch cohorts. Cell Rep. 38, 110204 (2022).
Google Scholar
Yang, K. et al. Alterations in the gut virome in obesity and type 2 diabetes mellitus. Gastroenterology 161, 1257–1269.e13 (2021).
Google Scholar
Clooney, A. G. et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26, 764–778.e5 (2019).
Google Scholar
Castellarin, M. et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012).
Google Scholar
Haghi, F., Goli, E., Mirzaei, B. & Zeighami, H. The association between fecal enterotoxigenic B. fragilis with colorectal cancer. BMC Cancer 19, 879 (2019).
Google Scholar
Bucher-Johannessen, C. et al. Long-term follow-up of colorectal cancer screening attendees identifies differences in Phascolarctobacterium spp. using 16S rRNA and metagenome sequencing. Front. Oncol. 13, 1183039 (2023).
Google Scholar
Scott, A. J. et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 68, 1624–1632 (2019).
Google Scholar
Hannigan, G. D., Duhaime, M. B., Ruffin, M. T. 4th, Koumpouras, C. C. & Schloss, P. D. Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio 9, e02248–18 (2018).
Google Scholar
Navarro, M., Nicolas, A., Ferrandez, A. & Lanas, A. Colorectal cancer population screening programs worldwide in 2016: an update. World J. Gastroenterol. 23, 3632 (2017).
Google Scholar
Allison, J. E., Fraser, C. G., Halloran, S. P. & Young, G. P. Population screening for colorectal cancer means getting FIT: the past, present, and future of colorectal cancer screening using the fecal immunochemical test for hemoglobin (FIT). Gut Liver 8, 117–130 (2014).
Google Scholar
Rounge, T. B. et al. Evaluating gut microbiota profiles from archived fecal samples. BMC Gastroenterol. 18, 171 (2018).
Google Scholar
Krigul, K. L., Aasmets, O., Lüll, K., Org, T. & Org, E. Using fecal immunochemical tubes for the analysis of the gut microbiome has the potential to improve colorectal cancer screening. Sci. Rep. 11, 19603 (2021).
Google Scholar
Birkeland, E. et al. Profiling small RNAs in fecal immunochemical tests: is it possible? Mol. Cancer 22, 161 (2023).
Google Scholar
Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678 (2019).
Google Scholar
Van Doorslaer, K. et al. The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res. 45, D499–D506 (2017).
Google Scholar
Lopez, J. K. M. et al. Genomes of bacteriophages belonging to the orders Caudovirales and Petitvirales identified in fecal samples from Pacific flying fox (Pteropus tonganus) from the kingdom of Tonga. Microbiol. Resour. Announc. 11, e00038–22 (2022).
Google Scholar
Jansen, D. et al. Community types of the human gut virome are associated with endoscopic outcome in ulcerative colitis. J. Crohns Colitis 17, 1504–1513 (2023).
Google Scholar
Tang, Q. et al. Current sampling methods for gut microbiota: a call for more precise devices. Front. Cell. Infect. Microbiol. 10, 151 (2020).
Google Scholar
Gudra, D. et al. A widely used sampling device in colorectal cancer screening programmes allows for large-scale microbiome studies. Gut 68, 1723–1725 (2019).
Google Scholar
Masi, A. C. et al. Using faecal immunochemical test (FIT) undertaken in a national screening programme for large-scale gut microbiota analysis. Gut 70, 429–431 (2021).
Google Scholar
Bin Jang, H. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632–639 (2019).
Google Scholar
Minot, S. et al. Rapid evolution of the human gut virome. Proc. Natl Acad. Sci. USA 110, 12450–12455 (2013).
Google Scholar
Ramos-Barbero, M. D. et al. Characterization of crAss-like phage isolates highlights Crassvirales genetic heterogeneity and worldwide distribution. Nat. Commun. 14, 4295 (2023).
Google Scholar
Yutin, N. et al. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol. 3, 38–46 (2018).
Google Scholar
Zhang, M., Zhang, T., Yu, M., Chen, Y.-L. & Jin, M. The life cycle transitions of temperate phages: regulating factors and potential ecological implications. Viruses 14, 1904 (2022).
Google Scholar
Arnau, V. et al. Inference of the life cycle of environmental phages from genomic signature distances to their hosts. Viruses 15, 1196 (2023).
Google Scholar
Sutcliffe, S. G., Reyes, A. & Maurice, C. F. Bacteriophages playing nice: lysogenic bacteriophage replication stable in the human gut microbiota. iScience 26, 106007 (2023).
Google Scholar
Dikareva, E. et al. An extended catalog of integrated prophages in the infant and adult fecal microbiome shows high prevalence of lysogeny. Front. Microbiol. 14, 1254535 (2023).
Google Scholar
Luo, X.-Q. et al. Viral community-wide auxiliary metabolic genes differ by lifestyles, habitats, and hosts. Microbiome 10, 190 (2022).
Google Scholar
Asnicar, F. et al. Microbiome connections with host metabolism and habitual diet from 1098 deeply phenotyped individuals. Nat. Med. (2021).
Google Scholar
DeMarini, D. M. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat. Res. Mutat. Res. 567, 447–474 (2004).
Google Scholar
Joehanes, R. et al. Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 9, 436–447 (2016).
Google Scholar
Johansen, J. et al. Centenarians have a diverse gut virome with the potential to modulate metabolism and promote healthy lifespan. Nat. Microbiol. 8, 1064–1078 (2023).
Google Scholar
World Cancer Research Fund/American Institute for Cancer Research. In Diet, Nutrition, Physical Activity and Cancer: A Global Perspective dietandcancerreport.org (2018).
Murphy, J., Mahony, J., Ainsworth, S., Nauta, A. & van Sinderen, D. Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl. Environ. Microbiol. 79, 7547–7555 (2013).
Google Scholar
Schulfer, A. et al. Fecal viral community responses to high-fat diet in mice. mSphere (2020).
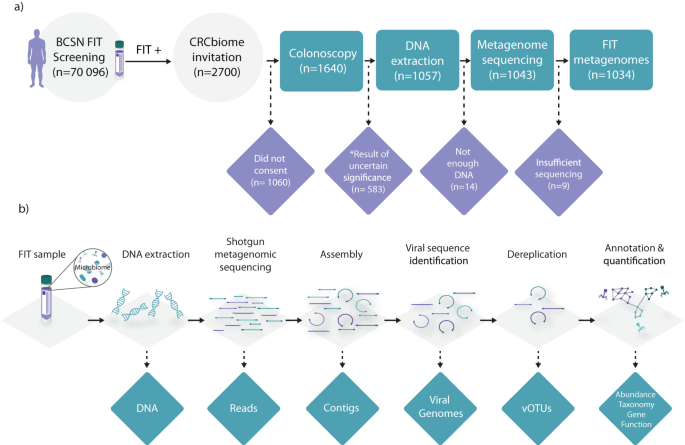
Kværner, A. S. et al. The CRCbiome study: a large prospective cohort study examining the role of lifestyle and the gut microbiome in colorectal cancer screening participants. BMC Cancer 21, 930 (2021).
Google Scholar
Xiao, L., Zhang, F. & Zhao, F. Large-scale microbiome data integration enables robust biomarker identification. Nat. Comput. Sci. 2, 307–316 (2022).
Google Scholar
Pardini, B. et al. A fecal MicroRNA signature by small RNA sequencing accurately distinguishes colorectal cancers: results from a multicenter study. Gastroenterology 165, 582–599.e8 (2023).
Google Scholar
Brunvoll, S. H. et al. Validation of repeated self-reported n-3 PUFA intake using serum phospholipid fatty acids as a biomarker in breast cancer patients during treatment. Nutr. J. 17, 94 (2018).
Google Scholar
Carlsen, M. H. et al. Evaluation of energy and dietary intake estimates from a food frequency questionnaire using independent energy expenditure measurement and weighed food records. Nutr. J. 9, 37 (2010).
Google Scholar
Andersen, L. F. et al. Evaluation of three dietary assessment methods and serum biomarkers as measures of fruit and vegetable intake, using the method of triads. Br. J. Nutr. 93, 519–527 (2005).
Google Scholar
Matvaretabellen. https://www.matvaretabellen.no/.
Shams-White, M. M. et al. Operationalizing the 2018 World Cancer Research Fund/American Institute for Cancer Research (WCRF/AICR) Cancer Prevention Recommendations: a standardized scoring system. Nutrients 11, 1572 (2019).
Google Scholar
Shams-White, M. M. et al. Further guidance in implementing the standardized 2018 World Cancer Research Fund/American Institute for Cancer Research (WCRF/AICR) Score. Cancer Epidemiol. Biomark. Prev. 29, 889–894 (2020).
Google Scholar
Helsedirektoratet (The Norwegian Directorate of Health). Anbefalinger Om Kosthold, Ernæring Og Fysisk Aktivitet (Recommendations for Diet. Nutrition and Physical Activity). https://www.helsedirektoratet.no/rapporter/anbefalinger-om-kosthold-ernaering-og-fysisk-aktivitet (2014).
Global Recommendations on Physical Activity for Health (World Health Organization, Geneva, 2010).
Piercy, K. L. et al. The physical activity guidelines for Americans. JAMA 320, 2020–2028 (2018).
Google Scholar
Kværner, A. S. et al. Associations of the 2018 World Cancer Research Fund/American Institute of Cancer Research (WCRF/AICR) cancer prevention recommendations with stages of colorectal carcinogenesis. Cancer Med. 12, 14806–14819 (2023).
Google Scholar
Kieser, S., Brown, J., Zdobnov, E. M., Trajkovski, M. & McCue, L. A. ATLAS: a Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data. BMC Bioinform. 21, 257 (2020).
Google Scholar
Bushnell, B. BBMap: BBMap short read aligner, and other bioinformatic tools. SourceForge https://sourceforge.net/projects/bbmap/ (2022).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017).
Google Scholar
Guo, J. et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 9, 37 (2021).
Google Scholar
Nayfach, S. et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 39, 578–585 (2021).
Google Scholar
Woodcroft, B. J. Galah – More scalable dereplication for metagenome assembled genomes https://github.com/wwood/galah. (2023).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 119 (2010).
Google Scholar
Cook, R. et al. INfrastructure for a PHAge REference Database: identification of large-scale biases in the current collection of cultured phage genomes. PHAGE 2, 214–223 (2021).
Google Scholar
Mihara, T. et al. Linking virus genomes with host taxonomy. Viruses 8, 66 (2016).
Google Scholar
Pandolfo, M., Telatin, A., Lazzari, G., Adriaenssens, E. M. & Vitulo, N. MetaPhage: an automated pipeline for analyzing, annotating, and classifying bacteriophages in metagenomics sequencing data. mSystems 7, e00741–22 (2022).
Google Scholar
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Google Scholar
Shaffer, M. et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 48, 8883–8900 (2020).
Google Scholar
Mistry, J. et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021).
Google Scholar
Thannesberger, J. et al. Viruses comprise an extensive pool of mobile genetic elements in eukaryote cell cultures and human clinical samples. FASEB J. 31, 1987–2000 (2017).
Google Scholar
Aramaki, T. et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinform. Oxf. Engl. 36, 2251–2252 (2020).
Google Scholar
Zhang, H. et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101 (2018).
Google Scholar
Brister, J. R., Ako-adjei, D., Bao, Y. & Blinkova, O. NCBI viral genomes resource. Nucleic Acids Res. 43, D571–D577 (2015).
Google Scholar
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Google Scholar
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLOS Comput. Biol. 17, e1009442 (2021).
Google Scholar
Olejnik, S. & Algina, J. Generalized Eta and Omega squared statistics: measures of effect size for some common research designs. Psychol. Methods 8, 434–447 (2003).
Google Scholar